The area I have chosen is on private land in a rural farm community outside of Taylor, British Columbia. It is approximately 12 acres in total with a predominately coniferous forest with a ravine that holds a small creek and both sides of the creek are farmed fields. The ravine itself boasts a lot of value with many different features such as small wetlands due to beaver (Castor canadensis) activity, pooling water and the running creek itself. It is important to note the creek is a lot more bank full than normal due to the increased amount of precipitation.
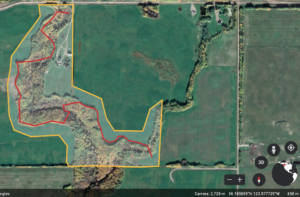
I first visited this area on July 14th, 2020 at 1400 hrs until approximately 1600 hrs. The weather was 20 degrees celsius with some overcast but, it was incredibly muggy. I remained in the area that is highlighted in yellow and scouted around within that perimeter that can be represented in the red line. The size of the chosen observation area is approximately 0.31 square kilometres.
The vegetation from the bottom of the ravine consisted of typical wetter vegetation species such as:
- Lady fern (Athyrium filix-femina)
- Meadow Horsetail (Equisetum pratense)
And then carrying up the slope gradient the vegetation has noticeable changes leading into species such as
- Prickly Wild Rose (Rose acicularis)
- Cows Parsnip (Heracleum maculatum)
- A few willow species (Salix spp)
- Saskatoon (Amelanchier alnofolia)
- Soopolallie (Shepherdia canadensis)
Leaving the ravine and going into the field there are a good mix such as
- Foxtail Barley (Hordeum jubatum)
- Timothy (Phleum prantense)
- Kentucky Bluegrass (Poa prantensis)
- Northern Brome (Bromus inermis)
- Alsike Clover (Trifolium hybridum)
- Alfalfa (Medicago sativa)
This crop farmed field is mostly Alfalfa with the other species making random appearances throughout the field. It would be good to do a proper walk though out the field as well.

There was also noted to be Black bear activity (Ursus americanus) due to several ant hills being dug up and some old marking in trees as well as tracks.

There is also a lot of ungulate activity in terms of tracks and droppings. Mostly White tailed deer (Odocoileus virginianus) and Moose (Alces alces). Elk (Cervus canadensis) are also known to frequent the area but, determining the difference in tracks is a little tricky.
Potential subjects and relevant questions
Question 1: Do the species of ungulates that frequent the upper field selectively graze? (I feel this is where I will go with my study)
Question 2: Despite being older and potentially no longer in use, how much is the old beaver dam impacting the area of the lower ravine?
Question 3: Is this relatively small area able to support a Black bear for a suitable territory or is it a series of areas frequented?

